Bioinformatics team publishes variant calling workflow in Molecular Biology and Evolution
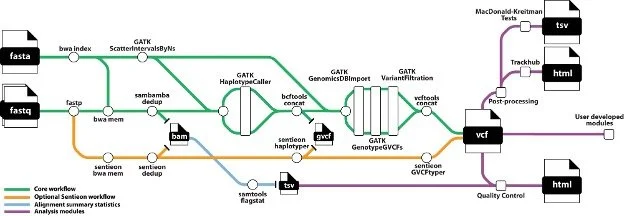
Figure 1. snpArcher overview. snpArcher is an automated pipeline implemented in Snakemake (Mölder et al. 2021). It takes short-read whole-genome sequencing data (fastq) and a reference genome as input and produces a multisample variant callset (VCF). With the modules presented here, snpArcher produces basic QC statistics and visualizations.
The CCGP bioinformatics team, led by past data wrangler Cade Mirchandani, and including current (Erik Enbody, Russ Corbett-Detig) and past (Mara Baylis) team members, along with several collaborators published the CCGP variant calling workflow in Molecular Biology and Evolution. The article entitled, “A fast, reproducible, high-throughput variant calling workflow for evolutionary, ecological, and conservation genomics” introduces snpArcher, a versatile and efficient workflow tailored for analyzing genomic resequencing data in non-model organisms. The authors demonstrate the workflow using 26 public resequencing datasets from non-mammalian vertebrates, showcasing its adaptability and potential for enabling comparative population genomic studies. This variant calling workflow is being used to process the resequencing data generated from thousands ofCCGP samples across well over 200 species. The paper describes a concise and innovative workflow to streamline the analysis of large genomic datasets, and contributes to enhancing our understanding of genetic variation across species.