Thomomys bottae (Pocket Gopher)
Visual overview of genome assembly metrics
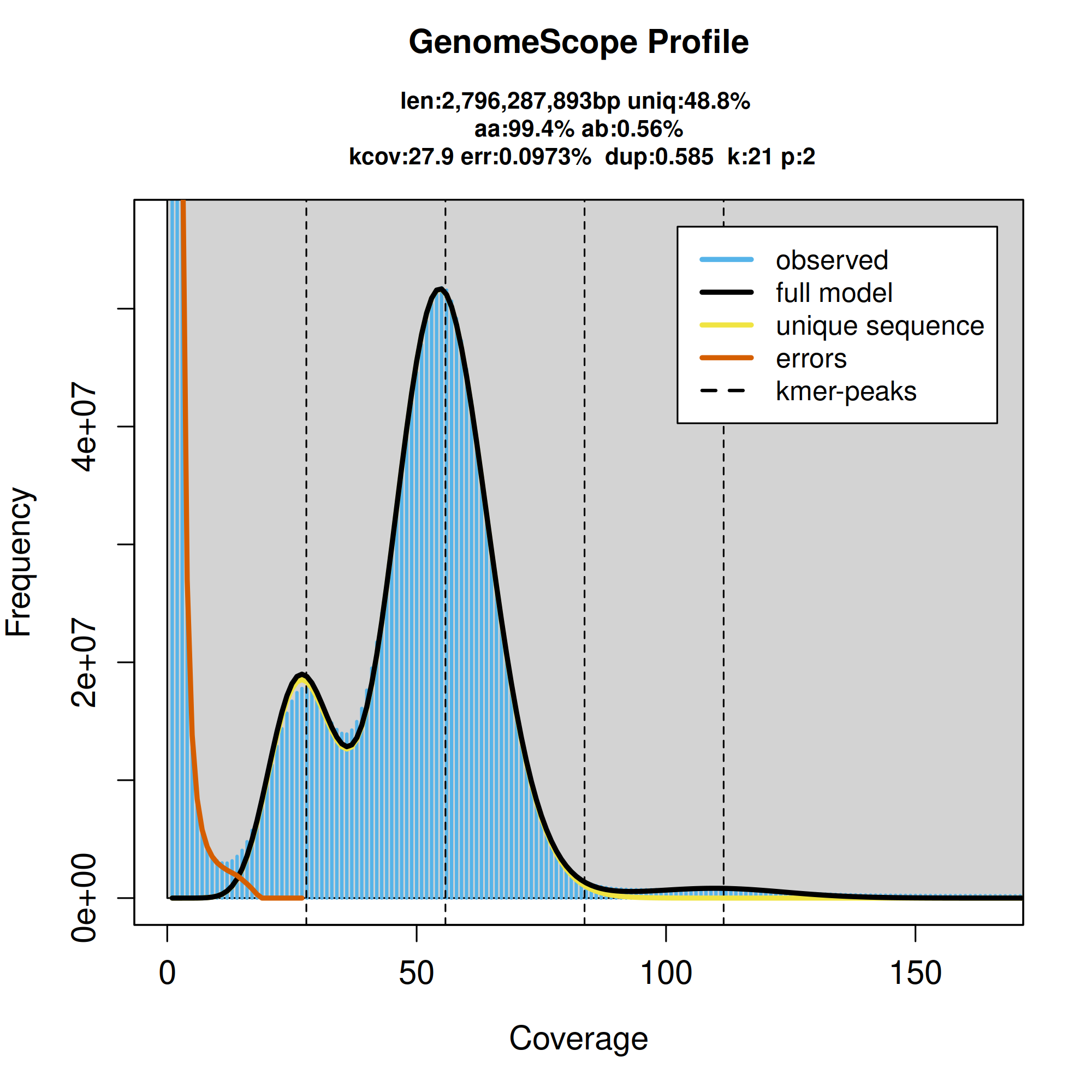
GenomeScope2.0 k-mer spectrum from adapter-trimmed HiFi sequence data. The bimodal pattern is expected for a diploid genome, with the left-hand peak representing k-mers in heterozygous regions and the right-hand peak representing k-mers in homozygous regions.
BlobToolKit Snail plot genome assembly summary. The central circle represents the length and number of scaffolds: the circumference of the circle represents the entire length of the genome and scaffolds are added in order of length starting from the longest scaffold to the shortest. The outer circle represents GC and AT content. BUSCO score is given in the upper right corner and length-related statistics are at the top left.
Contiguous linear organization of the primary and alternate genome assemblies visualized as an Omni-C contact map (PreTextSnapshot). Darker fill represents regions of the genome that are close to each other in 3D space as identified by chromatin-proximity sequencing. Black regions around the perimeter represent short fragments of the genome that could not be assembled into larger scaffolds. The large proportion of unassembled regions may refect high repeat content.
Authors
Erin R Voss, Merly Escalona, Krzysztof M Kozak, William Seligmann, Colin W Fairbairn, Oanh Nguyen, Mohan P A Marimuthu, Chris J Conroy, James L Patton, Rauri C K Bowie, Michael W Nachman